DeepMito
Mitochondrial proteins are physiologically active in different compartments and their aberrant localization contributes to the pathogenesis of human mitochondrial pathologies.
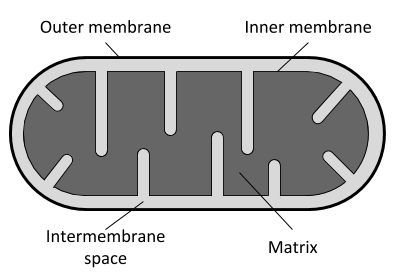
Here we present DeepMito, a novel method for predicting sub-mitochondrial localization and based on a convolutional neural network architecture to extract relevant patterns from primary features. Given an input protein, DeepMito is able to discriminate four different sub-mitochondrial compartments: outer membrane, inner membrane, intermembrane space and mitochondrial matrix.
More details on DeepMito can be found in the following paper:
Savojardo C, Bruciaferri N, Tartari G, Martelli PL, Casadio R. DeepMito: accurate prediction of protein submitochondrial localization using convolutional neural networks. Bioinformatics. 2019 Jun 20.