DeepMitoDB help
Basic search and browsing
The DeepMitoDB homepage (accessible at http://busca.biocomp.unibo.it/deepmitodb/) is shown in Figure 1.
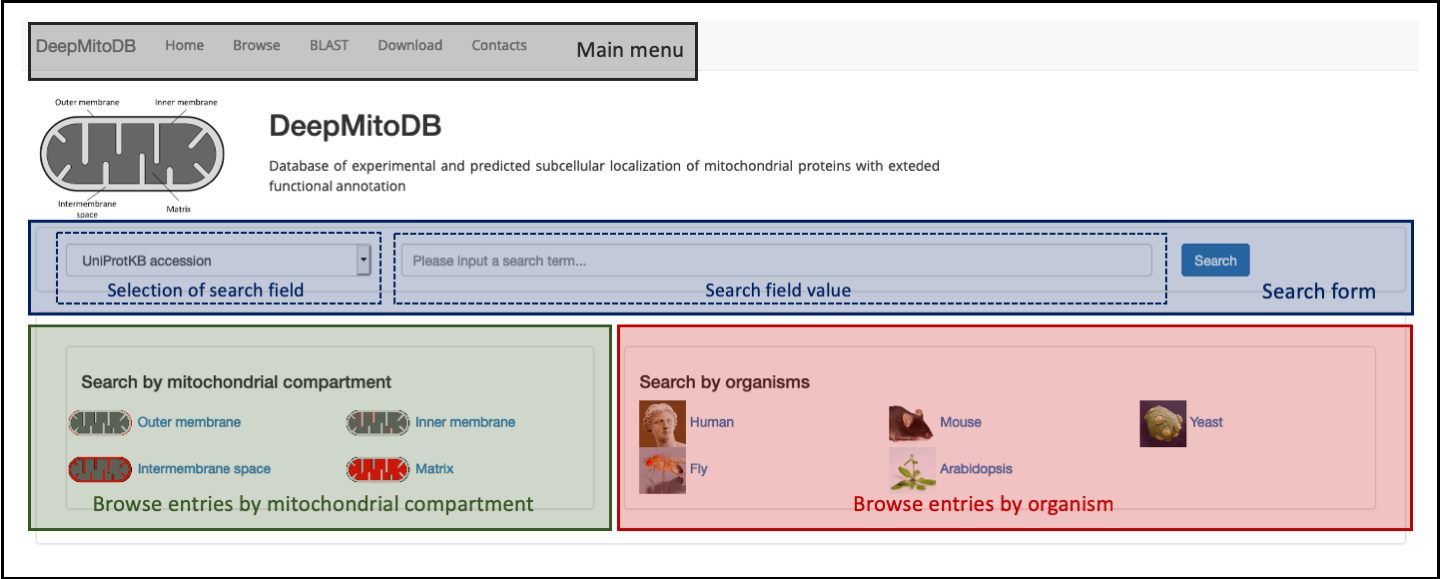
The homepage is organized into four different sections:
- The main menu (top of the page, highlighted in gray in Figure 1) provides links to the main website pages and it is shown in all pages during navigation.
- The search form (page center, highlighted in blue in Figure 1), allows users to search entries in tha database. Currently, users can choose between two different query types (via the select menu placed on the left side of the form): UniProKB accession (primary key of entries in DeepMitoDB) and Gene Ontology (GO) id. The latter search allows to obtain all entries in the database that are annotated with a specific GO term (in the molecular function or biological process aspect).
- Two panels (bottom of the page, highlighted in green and red in Figure 1), with links allowing to search by sub-mitochondrial compartment and organism, respectively.
The above described search options allow to restrict results to a subset of entries meeting specific criteria (accession, GO term, compartment or organism). Alternatively, the Browse (accessible from the main menu at http://busca.biocomp.unibo.it/deepmitodb/browse), allows to access all entries in the database. In both cases (i.e. restricted or unrestricted search), entries are listed as shown in Figure 2.
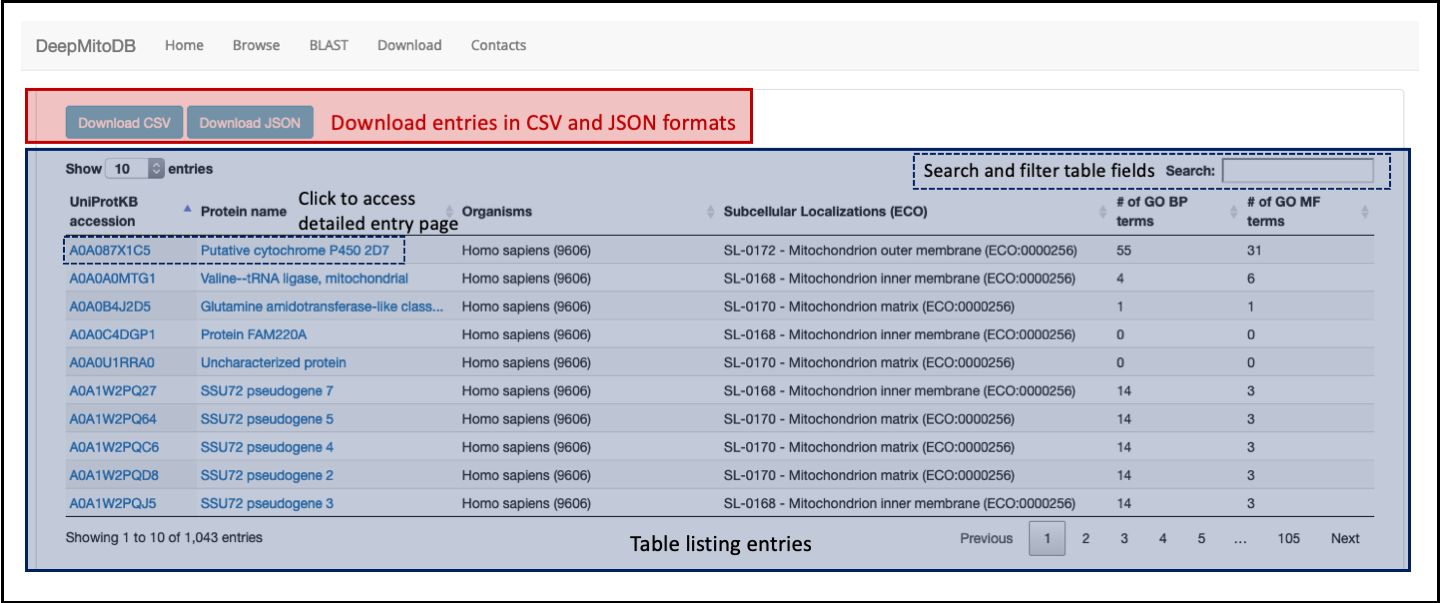
The main table (highlighted in blue in Figure 2) lists entries reporting, for each, the following information:
- The entry UniProtKB accession
- The protein name
- The protein source organism
- Sub-mitochondrial localizations (experimental or predicted), with the corresponding code from the Evidence and Conclusion Ontology (ECO)
- The number of biological process GO terms annotated on the protein
- The number of molecular function GO terms annotated on the protein
Entries listed in table can be searched and filtered using the search feature placed top-left of the main table. Clicking on the protein accession or name, users can access the detailed entry page (see next section). All data can be dowloaded in JSON and CSV formats (using the two buttons on top of tha table, highlighted in red in Figure 2).
The entry page
The DeepMitDB datailed entry page looks like the example (accessible here) shown in Figure 3.
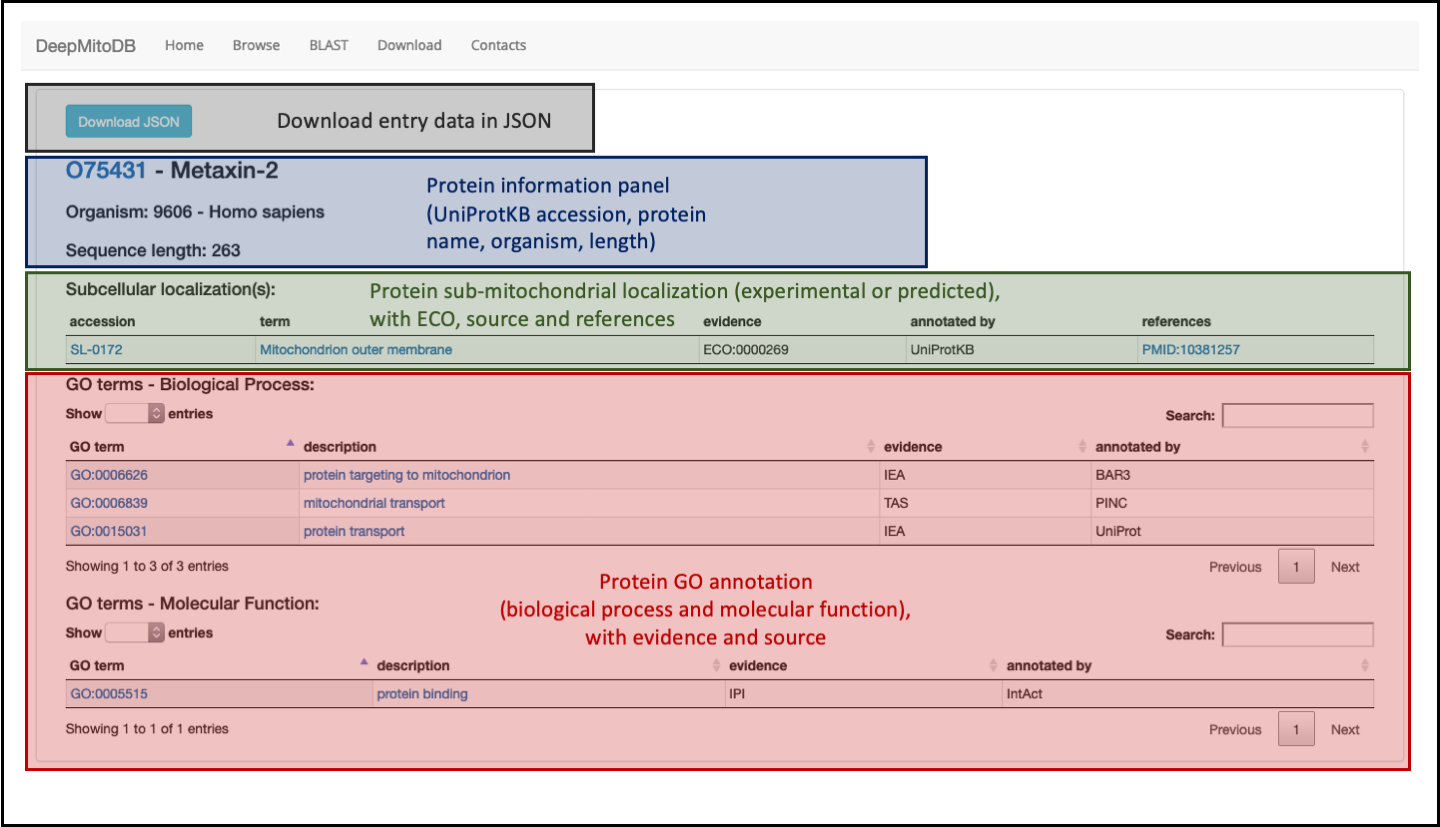
The page comprises three different sections:
- A panel providing general information about the protein entry such as UniProtKB accession, the protein name, the source organisms and the protein sequence length (blue panel in Figure 3).
- A table (in green in Figure 3) providing information about protein sub-mitochondrial localization and reporting, for each annotated localization, the accession (as reported in the UniProtDB Subcellular Location controlled vocabulary), the localization term, the annotation evidence code (ECO:0000269 or ECO:0000256 for experimental or predicted annotations, respectively), the annotation source and the supporting references (only for experimental annotations).
- Two additional tables (highlighted in red in Figure 3) listing annotated GO terms in the biological process and molecular function aspects. For each, the tables list the GO id, the GO term, the GO evidence code and annotation source
Entry data can be downloaded in JSON format (using the button in the grey section on top of the page in Figure 3).
BLAST search
The BLAST search page allows users to upload their own sequences and search for related proteins in DeepMitoDB. The input form is shhowed in Figure 4.
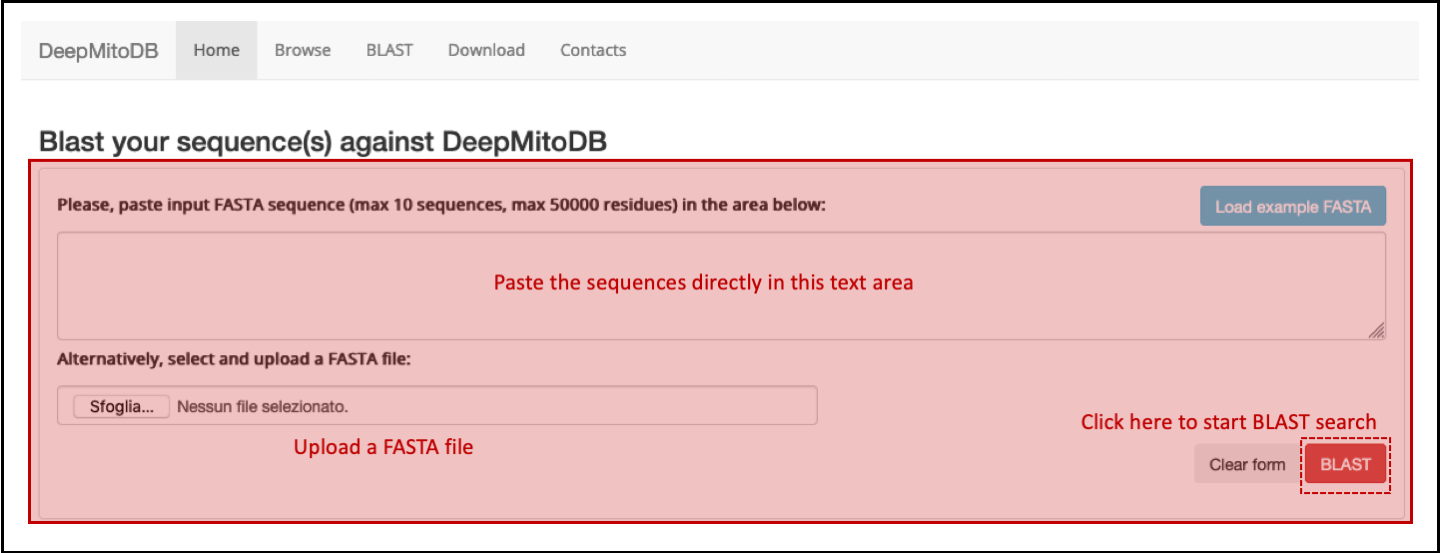
Up to 10 FASTA sequences can be pasted in text area or uploaded as external file. The BLAST search job can be then submitted to the server clicking the BLAST button. After job completion, results are listed as shown in Figure 5.
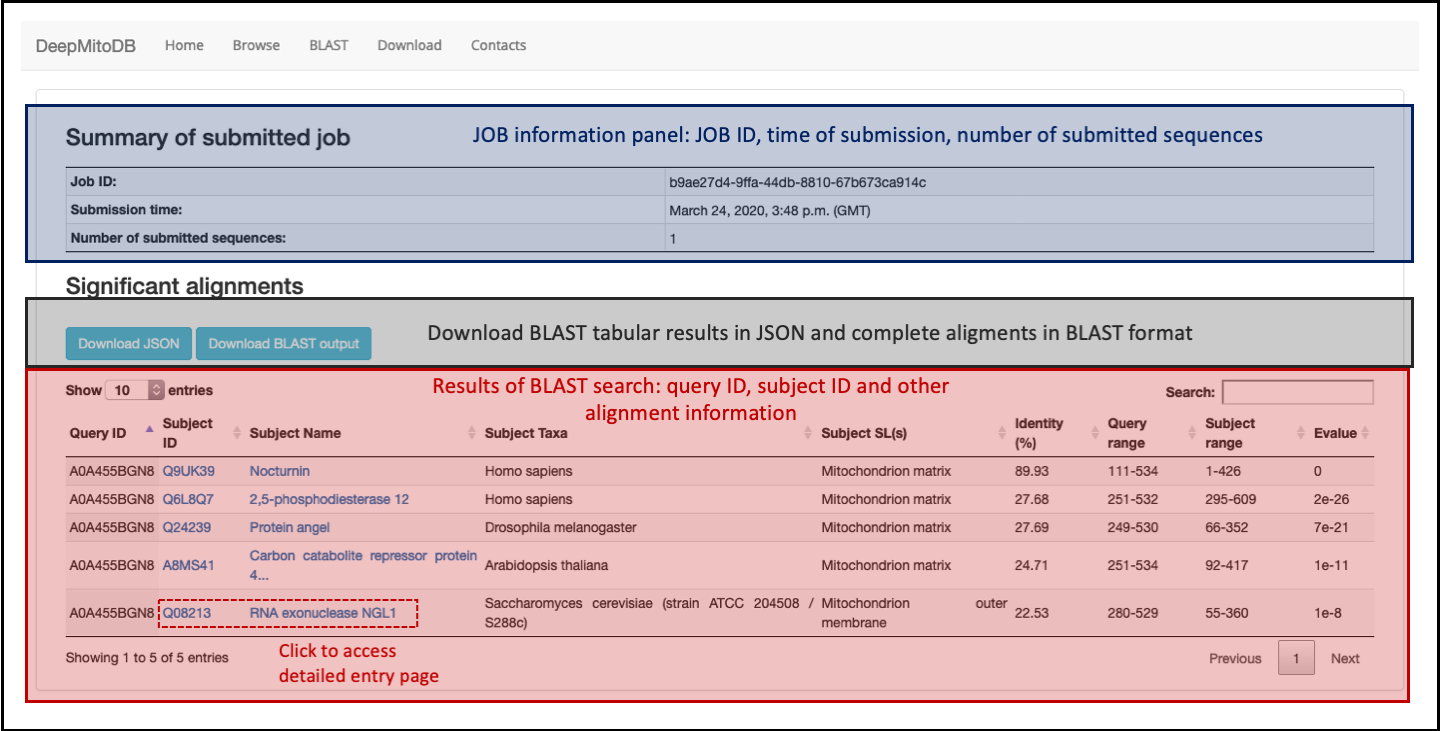
The BLAST result page comprises two sections:
- The top panel (in blue in Figure 5), reports summary information about the submitted job including: JOB ID, submission time and number of submitted sequences.
- The result table (in red in Figure 5), reporting significant alignments found. For each BLAST hit in the set of sequences inlcuded in DeepMitoDB, alignment results are listed, including: the query sequence ID, the subject sequence ID and name, the subject sequence source organism, annotated subcellular localization(s) of the subject sequence and alignment scores (sequence identity (%), query start-end residues, subject start-end residues and alignment e-value).
Users can access the detailed entry pages for each hit by clicking on the subject accession or name. Completed jobs can accessed via the provided JOB id (e.g. b9ae27d4-9ffa-44db-8810-67b673ca914c) at:
http://busca.biocomp.unibo.it/deepmitodb/<JOB-ID>/showresult/
JOB results will be mantained for a limited time (10 days).