The BetAware prediction server
BetAware is a web server for TransMembrane β-barrels detection and tooplogy prediction. Both prediction steps are based on advanced machine-learning methods.
Detection of β-barrels
TransMembrane β-barrels (TMBBs) are extremely important proteins that play key roles in several cell functions. They cross the lipid bilayer with β-barrels structures. TMBBs are presently found in the outer membranes of Gram-negative bacteria or in the membranes of mitochondria and chloroplasts. The loop exposure outside the cell membranes makes TMBBs important targets for vaccine or drug therapies. In genomes, they are not highly represented and are difficult to identify with experimental approaches. For these reasons several computational methods have been developed to discriminate them from other types of proteins. However, the best performing approaches have a high fraction of false positive predictions.
BetAware exploits a new machine learning approach based on N-to-1 Extreme Learning Machines[1] that significantly outperforms the previous methods achieving a Matthews correlation coefficient of 0.86, a probability of correct TMBB prediction of 1.0 and a sensitivity of 0.75.
TMBB topology prediction
TMBB topology prediction is an important step of TMBB protein structure prediction. Indeed, topologcial information about membrane-spanning beta-strands, inner and outer loops can be used to build a first, coarse model of a target protein.
BetAware predicts TMBB topology using a probabilistic model based on Grammatical-Restrained Hidden Conditional Random Fields, a discriminative framework introduced to address sequence labelling tasks in Bioinformatics[2].
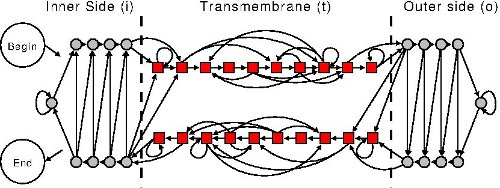
The topological model adopted by BetAware is depicted in Figure 1 as a Finite-State Machine which model membrane-spanning β-strands, inner and outer loops by incorporating several β-barrel construction rules and experimental segment length distributions. The model was trained using high-resolution TMBB structures taken from the Protein Data Bank.
References
- [1] Savojardo C., Fariselli P., Casadio R., Improving the detection of transmembrane β-barrel chains with N-to-1 Extreme Learning Machines, Bioinformatics (2011) 27 (22): 3123-3128.
- [2] Fariselli P., Savojardo C., Martelli P.L., Casadio R., Grammatical-Restrained Hidden Conditional Random Fields for Bioinformatics Applications, Algorithms for Molecular Biology 2009, 4:13.