Usage
The server accepts 1 protein sequences per job. The user can either paste the sequence in the text-area provided, or, alternatively, upload a FASTA file containing protein sequence.
The BetAware-Deep home page looks like the following:
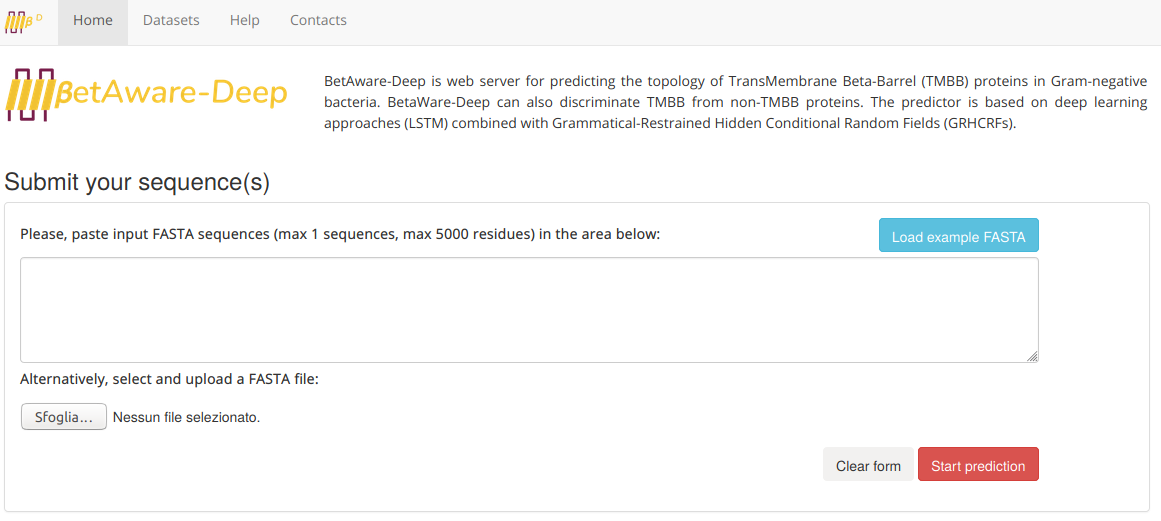
An example input sequence can be automatically loaded into the text-area using the "Example FASTA" button. To submit the job, the user should simply click on the "Start prediction" button.
Upon submission, the user will be automatically redirected to the page where the job results will be available after completion. If the job is still running, the result table shows no content.
Output
The BetAware-Deep output looks like the following:
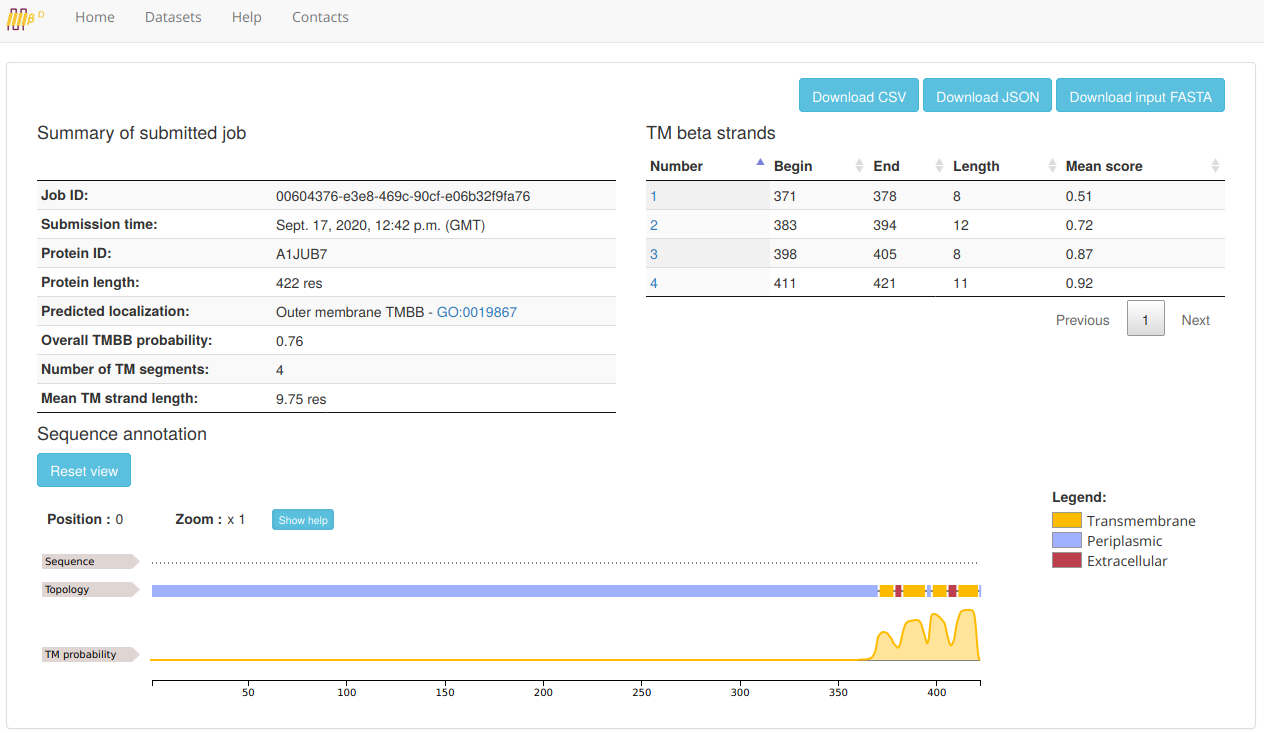
In the first section (the Summary table on the upper left side), main job information is reported.
Job information include:
- Job ID: a universal unique identifier which can be used to unambigously retrieve the job results after completion.
- Submission time: the GMT time of submission
- The protein ID/accession
- The protein length: the taxonomy the sequences belong to, as specified by the user at submission time
- Predicted protein localization: can be Outer membrane TMBB - GO:0019867 when more than 3 TM segments are predicted or other (less than 4 predicted TM stands)
- The overall TMBB probability: the probability, for the input protein, to be a TMBB protein
- The number of predicted TM segments
- The mean TM segment length
In the second section (the TM beta strands table on the upper right side), detailed information about predicted TM segments is reported, including:
- TM segment sequential number (from N- to C-terminus)
- Segement start position along the sequence
- Segement end position along the sequence
- Segement length
- Segment average probability score
By clicking on the segment number the user can zoom in on the corresponding TM strand on the sequence annotation view (see below)
.The section "Sequence annotation" contains the actual topology prediction result, displayed as a feature viewer object:
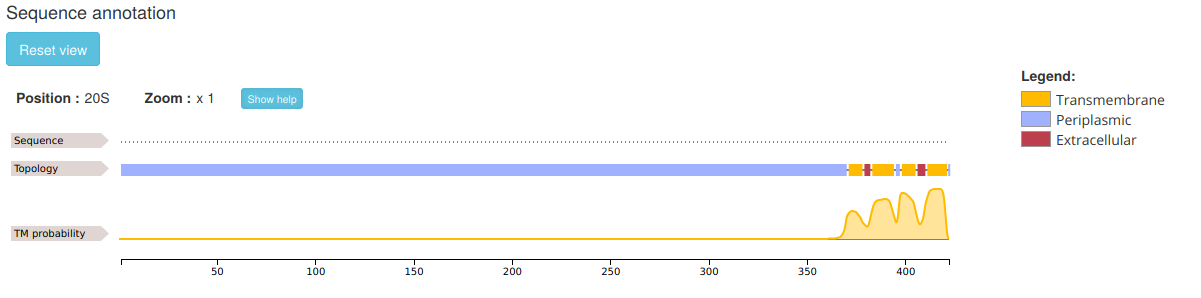
Two piaces of information are visualized on different tracks:
- Predicted TMBB topology, including protein segmentation in perisplasmic (blue), transmembrane (yellow) and extracellular (red).
- Residue-level TM probability score
The user can zoom in by left-clicking and selecting the area of interest. This allows to see feature annotation at residue-level:
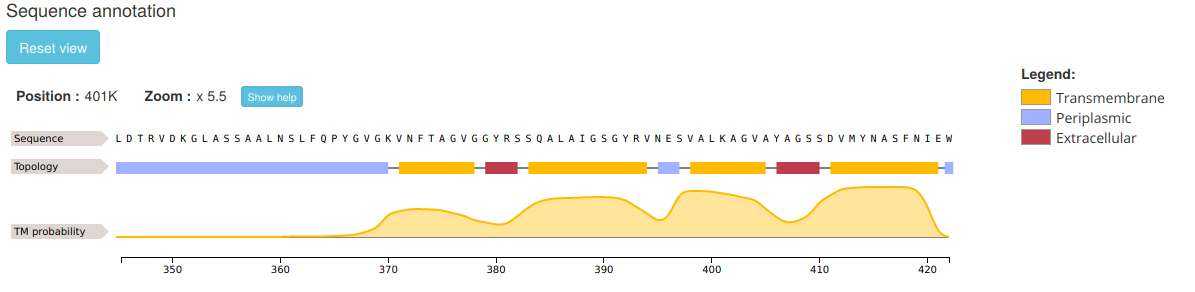
To zoom out just right-click on the feature track to reset the scale or press the Reset view button. Note that, sometimes, when the sequence is too long (as in the example shown), the actual residues are not visible at the initial default zoom level.
The button "Show help" provides an explaination on how to use the interactive feature panel.
Using the buttons "Download JSON", "Download CSV" and "Download input FASTA" the user can download the job results in JSON or CSV formats as well as the submitted protein sequence in FASTA format.