Help page
Submitting sequences to SVMyr
One or more protein sequences in FASTA format can be uploaded into the appropriate field (Figure 1). The server accepts at the most 5000 protein sequences. The user can either paste one or more sequences in the text-area provided, or, alternatively, upload a FASTA file containing protein sequences.
Before submission, the user can choose (using a checkbox) to perform prediction of:
- co-translational myristolation only
- post-translational myristoilation only
- both type of predictions at the same time.
In co-translational mode, SVMyr tries to identify myristolation sites
located at the N-termini of the input proteins. In this case, only proteins
starting either with a Glycine (G) or with a Methionine (M) followed by a G are considered for predictions.
Other proteins, not satisfying this condition are all predicted as non-myristoylated by default.
In post-translational mode, SVMyr attempts the protein-wide identification of myristolation sites.
In this case, caspase-cleavage sites are firstly identified by means of a combination of different regular expression
previously identified in literature. Then, myristolation prediction is perfored for those caspase-cleavage sites exposing a G at the N-terminus of the cleaved
peptide. When an input protein starts either with G or MG, also the predictio of the presence of an N-terminal myristolation sites is attempted.
Important note: post-translational myristoylation is relevant only when Metazoan sequences are submitted,
being this process observed so far only in Metazoa.
When post-translational prediction is included in the analysis, the user can decide whether adopting all caspase
cleavage-site motifs available at the Eukaryotic Linear Motif (ELM) database (Caspase 2, 6, 3-7 and 9), or restricting
the search to the Caspase 3-7 only, whose cleavage-site motif is the only one currently validated in ELM.
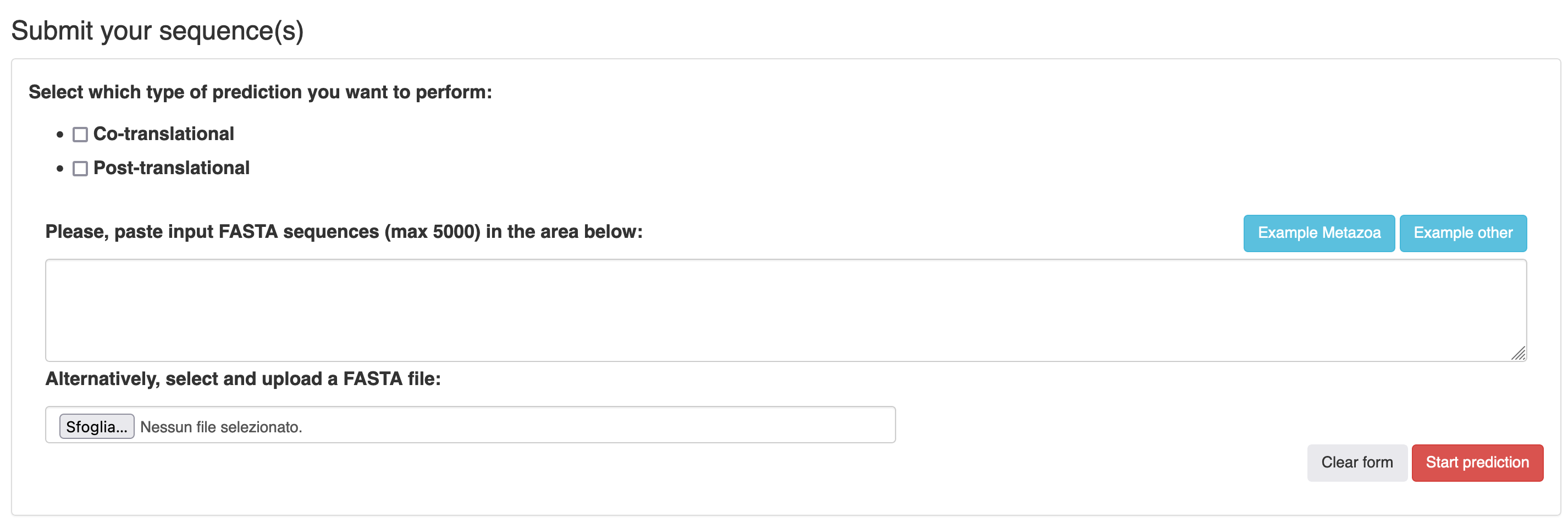
Figure 1. The SVMyr home page
Example input sequences can be automatically loaded into the text-area using the "Example Metazoa" and "Example other" buttons. The former loads an example job using Metazoan sequences, configuring the server to perform both co-translational and post-translational MYR predictions with all caspase motifs. The latter provides an example of sequences from non-metazoan eukaryotes to perform co-translational prediction only.
To submit the job, the user can simply click on the "Start prediction" button (Figure 1).
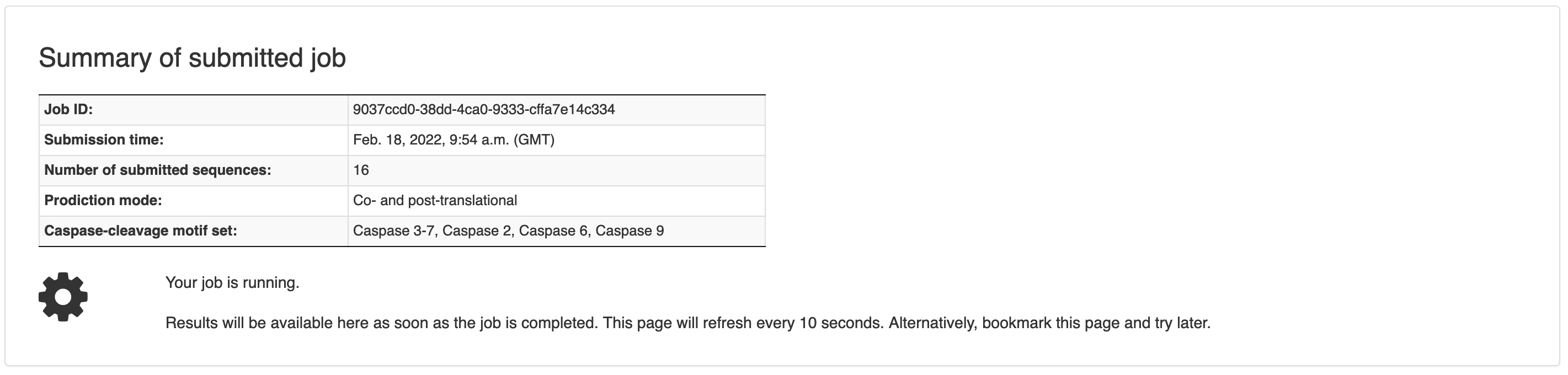
Figure 2. Job summary page
Upon submission, the user is automatically redirected to the page where the job results will be available after completion. If the job is still running, the result table shows no content (Figure 2). The page updates every 10 seconds and it will automatically show the results when ready
The SVMyr output page
An example of the main SVMyr output page is shown in Figure 3.
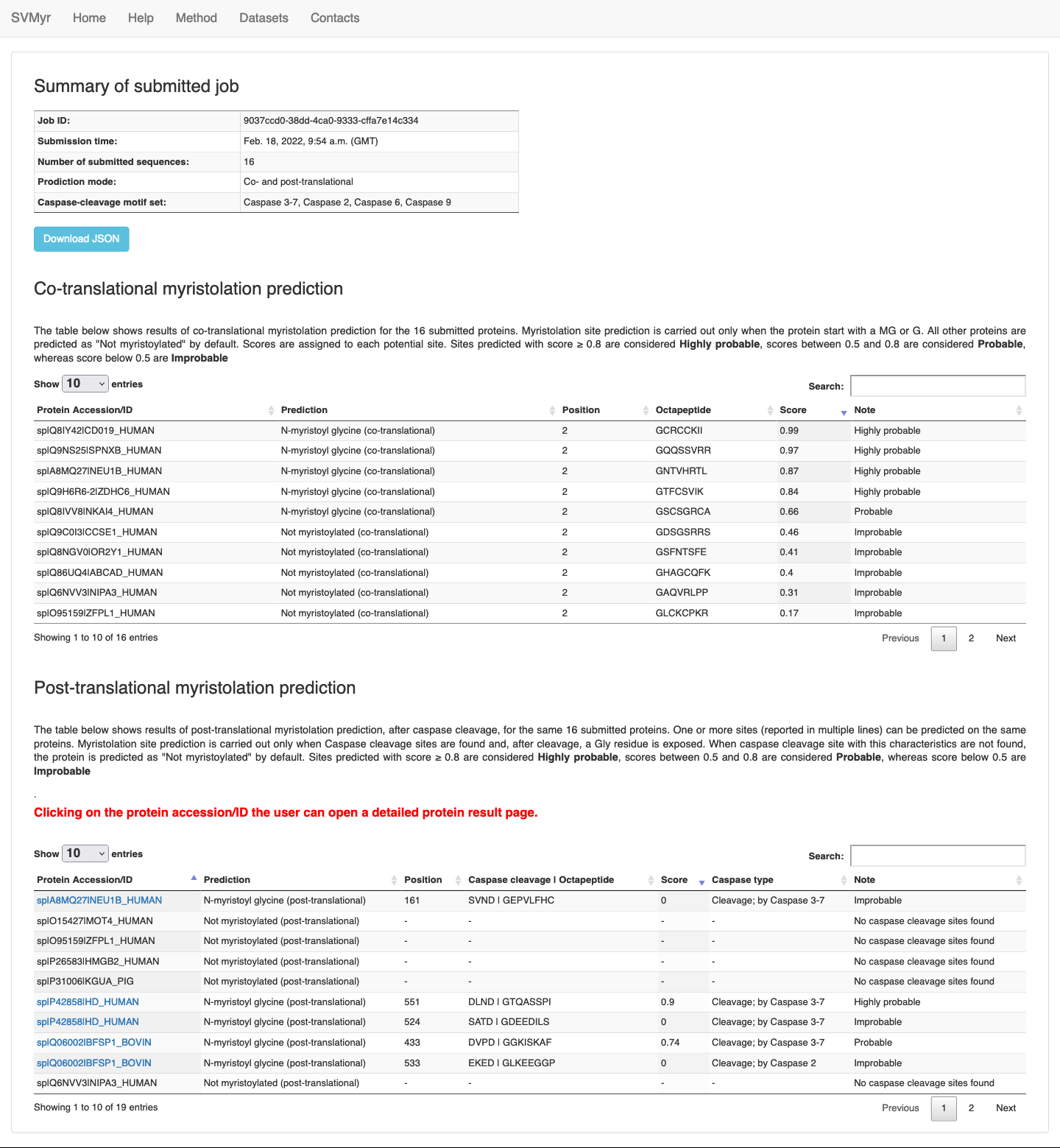
Figure 3. The output page
The table on top shows job information, which includes:
- Job ID: a universal unique identifier which can be used to unambigously retrieve the job results after completion.
- Submission time: the GMT time of submission
- The prediction type performed: co-translational, post-translational or both
- Number of submitted sequences: the total number of sequences included in the submission (either pasted in the text-area or uploaded as a file)
- The set of caspase cleavage-site motif adopted: all motifs (Caspase 2, 6, 3-7 and 9) or Caspase 3-7 only.
Using the button "Download JSON" the user can download the job complete results in JSON format (Figure 3).
The section "Co-translational myristoylation prediction" contains the prediction results for the identification of N-terminal myristoylation sites (when performed), displayed as a paginated table. By default, 10 entries are shown on each page. Clicking on the pagination buttons, the user can browse results. For each submitted protein, the following data are reported:
- the protein accession/ID (as can be read from the input FASTA)
- the myristoylation prediction: can be N-myristoyl glycine (co-translational) or Not myristoylated (co-translational)
- the position of the myristoylation site (not reported for non-myristoylated proteins)
- the octapeptide
- a prediction score, in the range [0-1]: sites predicted with score ≥ 0.8 are considered Highly probable; score between 0.5 and 0.8 are considered Probable, whereas score below 0.5 are Improbable.
The section "Post-translational myristoylation prediction" contains the prediction results for the identification of internal myristoylation sites (when performed), displayed as a paginated table. By default, 10 entries are shown on each page. Clicking on the pagination buttons, the user can browse results. For each submitted protein, the following data are reported:
- the protein accession/ID (as can be read from the input FASTA)
- the myristoylation prediction: can be N-myristoyl glycine (post-translational) or Not myristoylated (post-translational) (when no internal sites are found)
- the position of the myristoylation site (not reported for non-myristoylated proteins)
- the caspase cleavage match sequence | the myristoylation octapeptide
- The type of caspase cleavage sites found
- a prediction score, in the range [0-1]: sites predicted with score ≥ 0.8 are considered Highly probable; score between 0.5 and 0.8 are considered Probable, whereas score below 0.5 are Improbable.
Detailed protein results (post-translational prediction mode)
By clicking on the protein accession/ID, it is possible to visualize detailed results for the given specific protein, when available.
Different pieces of information are visualized.
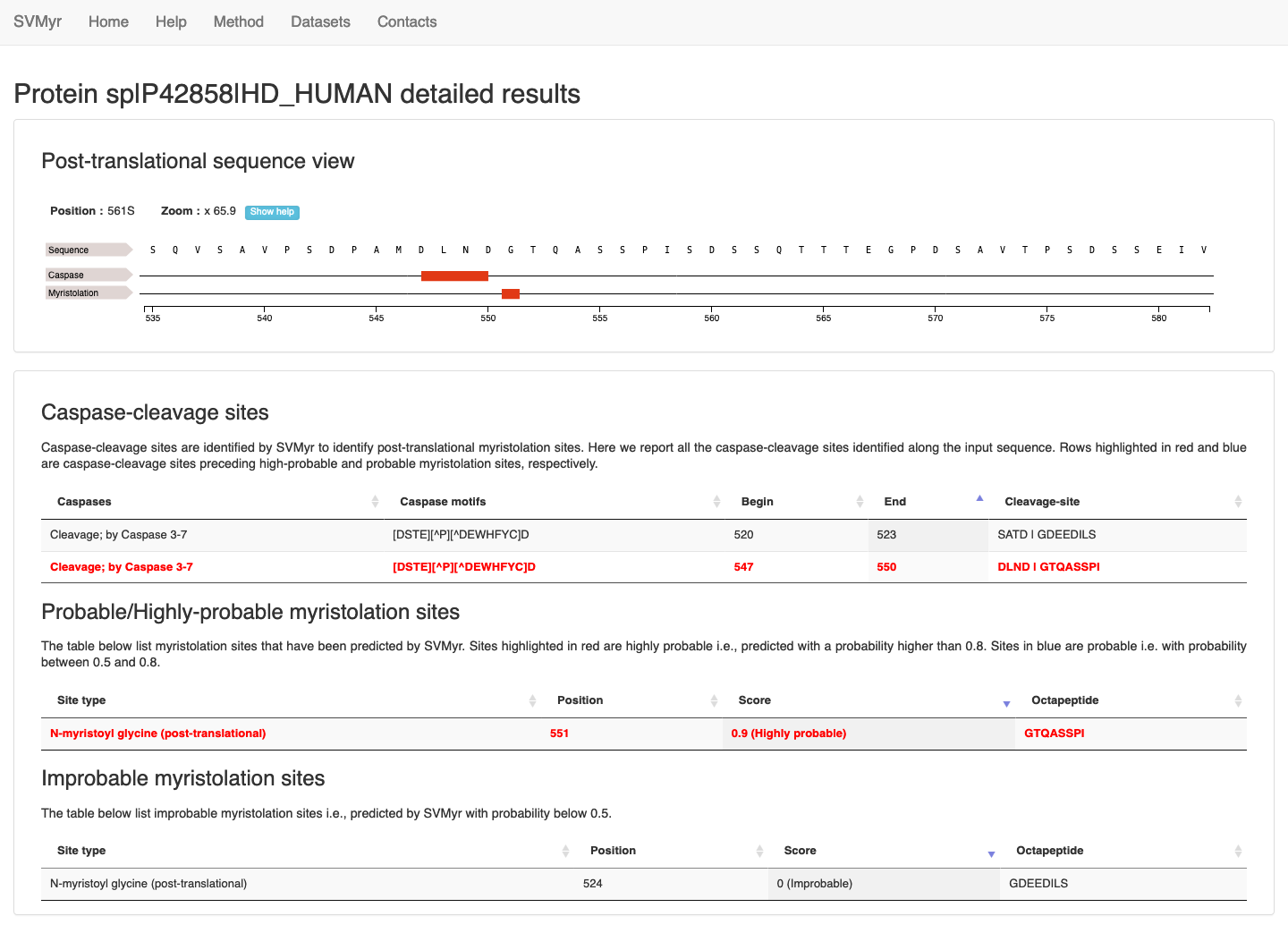
An interactive feature viever reports predicted caspase-cleavage and myristoylation sites (Figure 4). This allows to see feature annotation at residue-level.
The user can zoom in by left-clicking and selecting the area of interest. To zoom out just right-click on the feature track to reset the scale. Note that, sometimes, when the sequence is too long the actual residues are not visible at the initial default zoom level.
Three additional tables are also provided (Figure 4), showing detailed information about:
- all caspase-cleavage sites predicted along the sequence
- probable/highly-probable predicted myristoylation sites
- other sites predicted with score < 0.5, respectively.