Methods
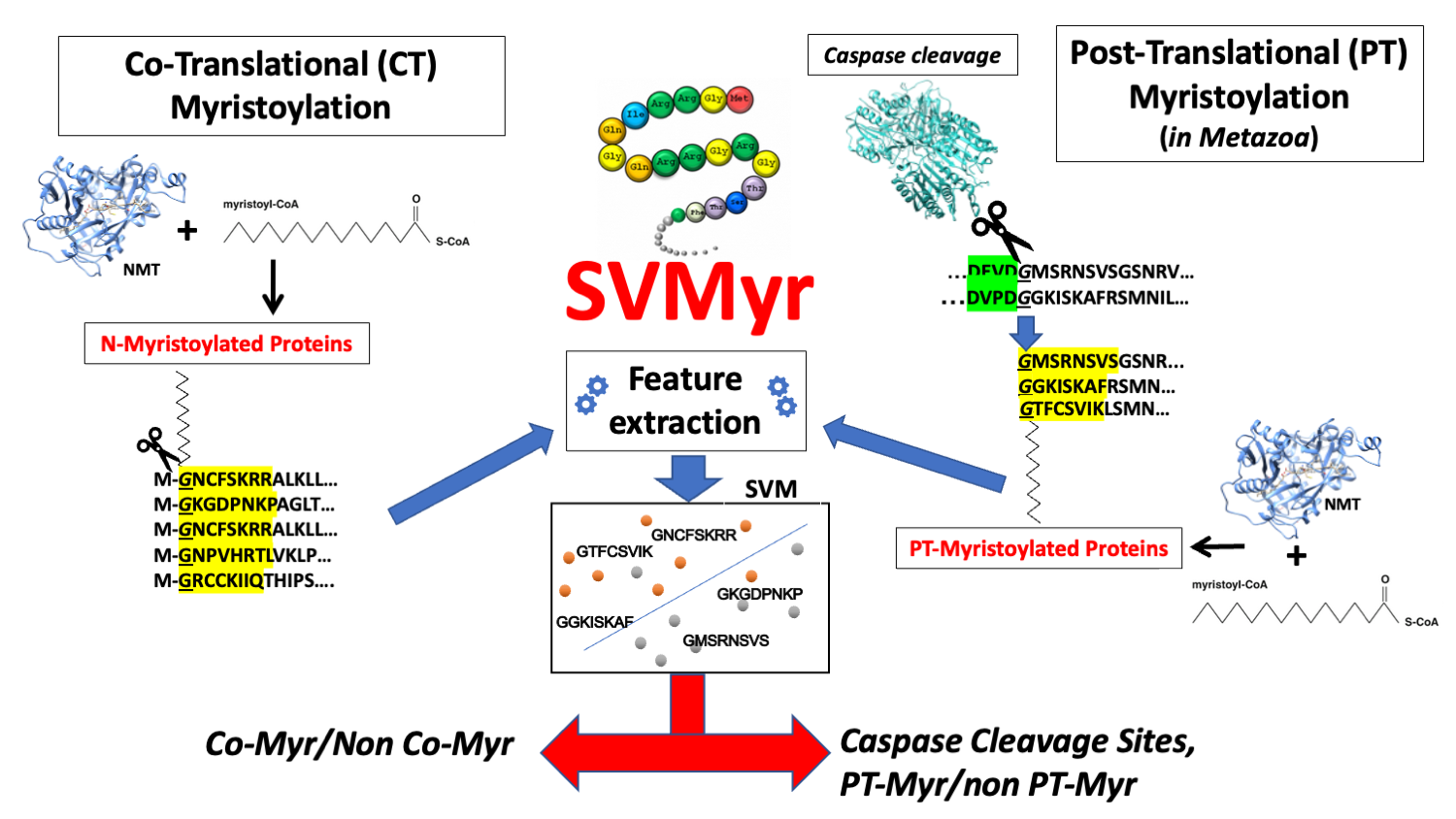
Figure 1. The SVMyr workflow
Input scheme
SVMyr (Figure 1) a Support Vector Machine (SVM)-based method tackling the problem of detecting both co-translational and, in metazoa, post-translational myristoylation sites.
Gly-octapeptide input includes seven scores towards a Position-Specific Scoring Matrix (PSSM), and average physicochemical characteristics (including hydrophobicity, charge, secondary structure propensity, and size).
PSSM is computed by stacking in a gapless mode all the Gly-octapeptides included in the positive training set of each cross-validation run. A profile is then computed as the frequency of the 20 residue types in each column in the alignment. A second, background profile to compute the log-odd scores in the PSSM, is obtained collecting all the N-terminal octapeptides of Gly or Met-Gly starting eukaryotic proteins included in SwissProt (14,304 non identical octapeptides).
The input Gly-octapeptide is then represented with seven positional scores (Gly is excluded) derived from PSSM, and with 5 physicochemical features averaged over the seven variable positions.
Physicochemical residue features include charge (considering charge +1 for Arg and Lys, and -1 for Asp and Glu), size as derived from AAindex [1] (https://www.genome.jp/aaindex), hydrophobicity according to the Kyte-Doolittle scale [2], and propensity towards alpha-helix and beta-strand secondary structures [3]. By this each Gly-octapeptide is encoded by a 12-dimensional vector.
SVM ensemble prediction and evaluation procedure
SVMyr predicts myristoylation sites by combining, with an ensemble majority scheme, the results of the 10 SVM models trained during the cross-validation procedure. Associated myristoylation probability is computed as the average of the output probabilities of the 10 SVMs in the ensemble and the input octapeptide is MYR predicted when the probability value is ≥ 0.50.
Prediction of post-translational MYR sites in Metazoa
SVMyr can search for internal myristoylation sites occurring in metazoan organisms, in which the attachment of the myristic group takes place at N-terminal glycine residues of polypeptides generated upon proteolytic cleavage by caspase enzymes. Apoptotic caspases seem to be mainly involved in the cleavage of post-translational MYR proteins in metazoa. To retrieve caspase cleavage site motifs, we refer to the Eukaryotic Linear Motifs (ELM) database [4]. In this database, we found four apoptotic caspase cleavage site motifs: one validated motif (ELME000321), for caspases 3/7, and three motifs reported as candidates in ELM (http://elm.eu.org/elms/candidates), for caspase 2, 6 and 9. Gly-starting octapeptides identified with this procedure are then classified with the ensemble SVMyr procedure.
References
[1] Kawashima, S. (2000). AAindex: Amino Acid index database. Nucleic Acids Res., 28, 374-374.
[2] Kyte, J., & Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105-132.
[3] Chou, P.Y., Fasman, G.D. (1974). Prediction of protein conformation. Biochemistry, 113, 222-245.
[4] Kumar, M., Gouw, M., Michael, S., Sámano-Sánchez, H., Pancsa, R., Glavina, J., Diakogianni, A., Valverde, J. A., Bukirova, D., Čalyševa, J., Palopoli, N., Davey, N. E., Chemes, L.B. & Gibson, T. J. (2020). ELM—the eukaryotic linear motif resource in 2020. Nucleic Acids Res., 48, D296-D306.