SChloro
SChloro is a predictor of protein sub-chloroplastic localization adopting a hierarchical prediction architecture based on SVM, which internally predicts presence/absence of chloroplastic targeting signals (including thylakoid targeting) and membrane topology interaction types (multi-pass, single-pass, peripheral)
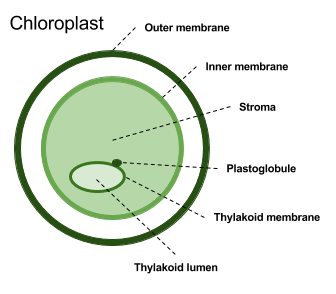
Overall, SChloro can discriminate 6 different chloroplastic compartments: Outer and inner membranes, stroma, plastoglobule, thylakoid membrane and lumen. When the proteins is predicted as having a tarnsit peptide, TPPred3 is adopted to identify the cleavage site. Moreover, transmembrane alpha helix are predicted using ENSEMBLE3.0.
More details on SChloro can be found in the following paper:
Savojardo C., Martelli P.L., Fariselli P., Casadio R. (2017) "SChloro: directing Viridiplantae proteins to six chloroplastic sub-compartments", Bioinformatics, 33(3), 347–353..